✅作者简介:热爱科研的Matlab仿真开发者,修心和技术同步精进,matlab项目合作可私信。
🍎个人主页:Matlab科研工作室
🍊个人信条:格物致知。
更多Matlab仿真内容点击👇
智能优化算法 神经网络预测 雷达通信 无线传感器
信号处理 图像处理 路径规划 元胞自动机 无人机 电力系统
⛄ 内容介绍
SPOD() 是频域形式的本征正交分解(POD,也称为主成分分析或 Karhunen-Loève 分解)的 Matlab 实现,称为谱本征正交分解 (SPOD)。SPOD 源自固定流 [1,2] 的时空 POD 问题,并导致每个模式都以单一频率振荡。SPOD 模式代表动态结构,可以最佳地解释平稳随机过程的统计变异性。
⛄ 部分代码
function [L,P,f,Lc,A] = spod(X,varargin)
%SPOD Spectral proper orthogonal decomposition
% [L,P,F] = SPOD(X) returns the spectral proper orthogonal decomposition
% of the data matrix X whose first dimension is time. X can have any
% number of additional spatial dimensions or variable indices. The
% columns of L contain the modal energy spectra. P contains the SPOD
% modes whose spatial dimensions are identical to those of X. The first
% index of P is the frequency and the last one the mode number ranked in
% descending order by modal energy. F is the frequency vector. If DT is
% not specified, a unit frequency sampling is assumed. For real-valued
% data, adjusted one-sided eigenvalue spectra are returned. Although
% SPOD(X) automatically chooses default spectral estimation parameters,
% the user is encouraged to manually specify problem-dependent parameters
% on a case-to-case basis.
%
% [L,P,F] = SPOD(X,WINDOW) uses a temporal window. If WINDOW is a vector,
% X is divided into segments of the same length as WINDOW. Each segment
% is then weighted (pointwise multiplied) by WINDOW. If WINDOW is a
% scalar, a Hamming window of length WINDOW is used. If WINDOW is omitted
% or set as empty, a Hamming window is used. Multitaper-Welch estimates
% are computed with the syntax SPOD(X,[NFFT BW],...), where WINDOW is an
% array of two scalars. NFFT is the window length and BW the
% time-halfbandwidth product. See [4] for details. By default, a number
% of FLOOR(2*BW)-1 discrete prolate spheroidal sequences (DPSS) is used
% as tapers. Custom tapers can be specified in a column matrix of tapers
% as WINDOW.
%
% [L,P,F] = SPOD(X,WINDOW,WEIGHT) uses a spatial inner product weight in
% which the SPOD modes are optimally ranked and orthogonal at each
% frequency. WEIGHT must have the same spatial dimensions as X.
%
% [L,P,F] = SPOD(X,WINDOW,WEIGHT,NOVERLAP) increases the number of
% segments by overlapping consecutive blocks by NOVERLAP snapshots.
% NOVERLAP defaults to 50% of the length of WINDOW if not specified.
%
% [L,P,F] = SPOD(X,WINDOW,WEIGHT,NOVERLAP,DT) uses the time step DT
% between consecutive snapshots to determine a physical frequency F.
%
% [L,P,F] = SPOD(XFUN,...,OPTS) accepts a function handle XFUN that
% provides the i-th snapshot as x(i) = XFUN(i). Like the data matrix X,
% x(i) can have any dimension. It is recommended to specify the total
% number of snaphots in OPTS.nt (see below). If not specified, OPTS.nt
% defaults to 10000. OPTS.isreal should be specified if a two-sided
% spectrum is desired even though the data is real-valued, or if the data
% is initially real-valued, but complex-valued for later snaphots.
%
% [L,P,F] = SPOD(X,WINDOW,WEIGHT,NOVERLAP,DT,OPTS) specifies options:
% OPTS.savefft: save FFT blocks to avoid storing all data in memory [{false} | true]
% OPTS.deletefft: delete FFT blocks after calculation is completed [{true} | false]
% OPTS.savedir: directory where FFT blocks and results are saved [ string | {'results'}]
% OPTS.savefreqs: save results for specified frequencies only [ vector | {all} ]
% OPTS.loadfft: load previously saved FFT blocks instead of recalculating [{false} | true]
% OPTS.mean: provide a mean that is subtracted from each snapshot [ array of size X | 'blockwise' | {temporal mean of X; 0 if XFUN} ]
% OPTS.nsave: number of most energtic modes to be saved [ integer | {all} ]
% OPTS.isreal: complex-valuedity of X or represented by XFUN [{determined from X or first snapshot if XFUN is used} | logical ]
% OPTS.nt: number of snapshots [ integer | {determined from X; defaults to 10000 if XFUN is used}]
% OPTS.conflvl: confidence interval level [ scalar between 0 and 1 | {0.95} ]
% OPTS.normvar: normalize each block by pointwise variance [{false} | true]
%
% [L,PFUN,F] = SPOD(...,OPTS) returns a function PFUN instead of the SPOD
% data matrix P if OPTS.savefft is true. The function returns the j-th
% most energetic SPOD mode at the i-th frequency as p = PFUN(i,j) by
% reading the modes from the saved files. Saving the data on the hard
% Determine whether data is real-valued or complex-valued to decide on one-
% or two-sided spectrum. If "opts.isreal" is not set, determine from data.
% If data is provided through a function handle XFUN and opts.isreal is not
% specified, deternime complex-valuedity from first snapshot.
if isfield(opts,'isreal')
isrealx = opts.isreal;
elseif ~xfun
isrealx = isreal(X);
else
x1 = X(1);
isrealx = isreal(x1);
clear x1;
end
% get default spectral estimation parameters and options
[window,weight,nOvlp,dt,nDFT,nBlks,nTapers] = spod_parser(nt,nx,isrealx,varargin{:});
nSamples = nBlks*nTapers;
% determine correction for FFT window gain
winWeight = 1./mean(abs(window)); % row vector for multitaper
% Use data mean if not provided through "opts.mean".
blk_mean = false;
if isfield(opts,'mean')
if strcmp('blockwise',opts.mean)
blk_mean = true;
end
end
if blk_mean
mean_name = 'blockwise mean';
elseif isfield(opts,'mean')
x_mean = opts.mean(:);
mean_name = 'user specified';
else
switch xfun
case true
x_mean = 0;
warning('No mean subtracted. Consider providing long-time mean through "opts.mean" for better accuracy at low frequencies.');
mean_name = '0';
case false
x_mean = mean(X,1);
x_mean = x_mean(:);
mean_name = 'data mean';
end
end
disp(['Mean : ' mean_name]);
% obtain frequency axis
f = (0:nDFT-1)/dt/nDFT;
if isrealx
f = (0:ceil(nDFT/2))/nDFT/dt;
else
if mod(nDFT,2)==0
f(nDFT/2+1:end) = f(nDFT/2+1:end)-1/dt;
else
f((nDFT+1)/2+1:end) = f((nDFT+1)/2+1:end)-1/dt;
end
end
nFreq = length(f);
% set default for confidence interval
if nargout>=4
confint = true;
if ~isfield(opts,'conflvl')
opts.conflvl = 0.95;
end
xi2_upper = 2*gammaincinv(1-opts.conflvl,nBlks);
xi2_lower = 2*gammaincinv( opts.conflvl,nBlks);
Lc = zeros(nFreq,nBlks,2);
else
confint = false;
Lc = [];
end
% set defaults for options for saving FFT data blocks
if ~isfield(opts,'savefreqs'), opts.savefreqs = 1:nFreq; end
if opts.savefft
if ~isfield(opts,'savedir'), opts.savedir = pwd; end
saveDir = fullfile(opts.savedir,['nfft' num2str(nDFT) '_novlp' num2str(nOvlp) '_nblks' num2str(nSamples)]);
if ~exist(saveDir,'dir'), mkdir(saveDir); end
if ~isfield(opts,'nsave'), opts.nsave = nSamples; end
if ~isfield(opts,'deletefft'), opts.deletefft = true; end
omitFreqIdx = 1:nFreq; omitFreqIdx(opts.savefreqs) = [];
end
if ~isfield(opts,'loadfft'), opts.loadfft = false; end
if ~isfield(opts,'normvar'), opts.normvar = false; end
% loop over number of blocks and generate Fourier realizations
disp(' ')
disp('Calculating temporal DFT')
disp('------------------------------------')
if ~opts.savefft
Q_hat = zeros(nFreq,nx,nSamples);
end
Q_blk = zeros(nDFT,nx);
for iBlk = 1:nBlks
% check if all FFT files are pre-saved
all_exist = 0;
if opts.loadfft
for iFreq = opts.savefreqs
if ~isempty(dir(fullfile(saveDir,['fft_block' num2str([iBlk iFreq],'%.4i_freq%.4i.mat')])))
all_exist = all_exist + 1;
end
end
end
if opts.loadfft && all_exist==length(opts.savefreqs)
disp(['loading FFT of block ' num2str(iBlk) '/' num2str(nBlks) ' from file'])
else
% get time index for present block
offset = min((iBlk-1)*(nDFT-nOvlp)+nDFT,nt)-nDFT;
timeIdx = (1:nDFT) + offset;
% build present block
if blk_mean, x_mean = 0; end
if xfun
for ti = timeIdx
x = X(ti);
Q_blk(ti-offset,:) = x(:) - x_mean;
end
else
Q_blk = bsxfun(@minus,X(timeIdx,:),x_mean.');
end
% if block mean is to be subtracted, do it now that all data is
% collected
if blk_mean
Q_blk = bsxfun(@minus,Q_blk,mean(Q_blk,1));
end
% normalize by pointwise variance
if opts.normvar
Q_var = sum(bsxfun(@minus,Q_blk,mean(Q_blk,1)).^2,1)/(nDFT-1);
% address division-by-0 problem with NaNs
Q_var(Q_var<4*eps) = 1;
Q_blk = bsxfun(@rdivide,Q_blk,Q_var);
end
for iTaper = 1:nTapers
iSample = iBlk+((iTaper-1)*nBlks);
if nTapers>1
taperString = [', taper ' num2str(iTaper) '/' num2str(nTapers)];
else
taperString = [];
end
disp(['block ' num2str(iBlk) '/' num2str(nBlks) taperString ' (snapshots ' ...
num2str(timeIdx(1)) ':' num2str(timeIdx(end)) ')'])
% window and Fourier transform block
Q_blk_win = bsxfun(@times,Q_blk,window(:,iTaper));
Q_blk_hat = winWeight(iTaper)/nDFT*fft(Q_blk_win);
Q_blk_hat = Q_blk_hat(1:nFreq,:);
if ~opts.savefft
% keep FFT blocks in memory
Q_hat(:,:,iSample) = Q_blk_hat;
else
for iFreq = opts.savefreqs
file = fullfile(saveDir,['fft_block' num2str([iSample iFreq],'%.4i_freq%.4i')]);
Q_blk_hat_fi = single(Q_blk_hat(iFreq,:));
save(file,'Q_blk_hat_fi','-v7.3');
end
end
end
end
end
% loop over all frequencies and calculate SPOD
L = zeros(nFreq,nSamples);
A = zeros(nFreq,nSamples,nSamples);
disp(' ')
if nTapers>1
disp('Calculating Multitaper SPOD')
else
disp('Calculating SPOD')
end
disp('------------------------------------')
% unbiased estimator of CSD
if blk_mean
nIndep = nSamples-1;
else
nIndep = nSamples;
end
if ~opts.savefft
% keep everything in memory (default)
P = zeros(nFreq,nx,nSamples);
for iFreq = 1:nFreq
disp(['frequency ' num2str(iFreq) '/' num2str(nFreq) ' (f=' num2str(f(iFreq),'%.3g') ')'])
Q_hat_f = squeeze(Q_hat(iFreq,:,:));
M = Q_hat_f'*bsxfun(@times,Q_hat_f,weight)/nIndep;
[Theta,Lambda] = eig(M);
Lambda = diag(Lambda);
[Lambda,idx] = sort(Lambda,'descend');
Theta = Theta(:,idx);
Psi = Q_hat_f*Theta*diag(1./sqrt(Lambda)/sqrt(nIndep));
P(iFreq,:,:) = Psi;
A(iFreq,:,:) = diag(sqrt(nBlks)*sqrt(Lambda))*Theta';
L(iFreq,:) = abs(Lambda);
% adjust mode energies for one-sided spectrum
if isrealx
if iFreq~=1&&iFreq~=nFreq
L(iFreq,:) = 2*L(iFreq,:);
end
end
if confint
Lc(iFreq,:,1) = L(iFreq,:)*2*nIndep/xi2_lower;
Lc(iFreq,:,2) = L(iFreq,:)*2*nIndep/xi2_upper;
end
end
P = reshape(P,[nFreq dim(2:end) nSamples]);
else
% save FFT blocks on hard drive (for large data)
for iFreq = opts.savefreqs
disp(['frequency ' num2str(iFreq) '/' num2str(nFreq) ' (f=' num2str(f(iFreq),'%.3g') ')'])
% load FFT data from previously saved file
Q_hat_f = zeros(nx,nSamples);
for iBlk = 1:nSamples
file = fullfile(saveDir,['fft_block' num2str([iBlk iFreq],'%.4i_freq%.4i')]);
load(file,'Q_blk_hat_fi');
Q_hat_f(:,iBlk) = Q_blk_hat_fi;
end
M = Q_hat_f'*bsxfun(@times,Q_hat_f,weight)/nIndep;
[Theta,Lambda] = eig(M);
Lambda = diag(Lambda);
[Lambda,idx] = sort(Lambda,'descend');
Theta = Theta(:,idx);
Psi = Q_hat_f*Theta*diag(1./sqrt(Lambda)/sqrt(nIndep));
A(iFreq,:,:) = diag(sqrt(nIndep)*sqrt(Lambda))*Theta';
if opts.nsave>0
Psi = single(reshape(Psi(:,1:opts.nsave),[dim(2:end) opts.nsave]));
file = fullfile(saveDir,['spod_f' num2str(iFreq,'%.4i')]);
save(file,'Psi','-v7.3');
else
Psi = [];
end
L(iFreq,:) = abs(Lambda);
% adjust mode energies for one-sided spectrum
if isrealx
if iFreq~=1&&iFreq~=nFreq
L(iFreq,:) = 2*L(iFreq,:);
end
end
if confint
Lc(iFreq,:,1) = L(iFreq,:)*2*nIndep/xi2_lower;
Lc(iFreq,:,2) = L(iFreq,:)*2*nIndep/xi2_upper;
end
end
% return anonymous function that loads SPOD modes from files instead of
% actual solution
P = @(iFreq,iMode) getmode(iFreq,iMode,saveDir);
file = fullfile(saveDir,'spod_energy');
save(file,'L','Lc','f','A','-v7.3');
% clean up
if opts.deletefft
for iFreq = opts.savefreqs
for iBlk = 1:nSamples
file = fullfile(saveDir,['fft_block' num2str([iBlk iFreq],'%.4i_freq%.4i.mat')]);
delete(file);
end
end
end
disp(' ')
disp(['Results saved in folder ' saveDir])
end
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
function [P] = getmode(iFreq,iMode,saveDir)
%GETMODE Anonymous function that loads SPOD modes from files
file = matfile(fullfile(saveDir,['spod_f' num2str(iFreq,'%.4i')]));
dim = size(file.Psi);
for di = 1:length(dim)-1
idx{di} = 1:dim(di);
end
idx{di+1} = iMode;
P = file.Psi(idx{:});
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
function [window,weight,nOvlp,dt,nDFT,nBlks,nTapers] = spod_parser(nt,nx,isrealx,varargin)
%SPOD_PARSER Parser for SPOD parameters
% read input arguments from cell array
window = []; weight = []; nOvlp = []; dt = [];
nvarargin = length(varargin);
if nvarargin >= 1
window = varargin{1};
if nvarargin >= 2
weight = varargin{2};
if nvarargin >= 3
nOvlp = varargin{3};
if nvarargin >= 4
dt = varargin{4};
end
end
end
end
if size(window,2)>size(window,1) % ensure column matrix format
window = permute(window,[2 1]);
end
% check arguments and determine default spectral estimation parameters
% window size and type
if isempty(window)
nDFT = 2^floor(log2(nt/10));
window = hammwin(nDFT);
window_name = 'Hamming';
elseif length(window)==1
nDFT = window;
window = hammwin(window);
window_name = 'Hamming';
elseif length(window)==2
nDFT = window(1);
bw = window(2);
nTapers = floor(2*bw)-1;
window = slepsec(nDFT,bw,nTapers);
window_name = 'DPSS';
elseif length(window) == 2^nextpow2(length(window))
nDFT = length(window);
window_name = 'user specified';
else
nDFT = length(window);
window_name = 'user specified';
end
nTapers = size(window,2);
weight = weight(:);
% block overlap
if isempty(nOvlp)
nOvlp = floor(nDFT/2);
elseif nOvlp > nDFT-1
error('Overlap too large.')
end
% time step between consecutive snapshots
if isempty(dt)
dt = 1;
end
% inner product weight
if isempty(weight)
weight = ones(nx,1);
weight_name = 'uniform';
elseif numel(weight) ~= nx
error('Weights must have the same spatial dimensions as data.');
else
weight_name = 'user specified';
end
% number of blocks
nBlks = floor((nt-nOvlp)/(nDFT-nOvlp));
% test feasibility
if nDFT < 4 || nBlks*nTapers < 3
error('Spectral estimation parameters not meaningful.');
end
% display parameter summary
disp(' ')
if nTapers>1
disp('Multitaper SPOD parameters')
else
disp('SPOD parameters')
end
disp('------------------------------------')
if isrealx
disp('Spectrum type : one-sided (real-valued signal)')
else
disp('Spectrum type : two-sided (complex-valued signal)')
end
disp(['No. of snaphots per block : ' num2str(nDFT)])
disp(['Block overlap : ' num2str(nOvlp)])
disp(['No. of blocks : ' num2str(nBlks)])
disp(['Windowing fct. (time) : ' window_name])
disp(['Weighting fct. (space) : ' weight_name])
if nTapers>1
disp(['No. of tapers : ' num2str(nTapers)])
end
if exist('bw','var')
disp(['Time-halfbandwidth product: ' num2str(bw)])
end
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
function [window] = hammwin(N)
%HAMMWIN Standard Hamming window of lentgh N
window = 0.54-0.46*cos(2*pi*(0:N-1)/(N-1))';
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
function [window] = slepsec(nDFT,bw,nTapers)
%SLEPSEC Discrete prolate spheroidal sequences of length nDFT and
%time-halfbandwidth product bw
df = bw/nDFT;
j = 1:nDFT-1;
r1 = [df*2*pi, sin(2*pi*df*j)./j];
S = toeplitz(r1);
[U,L] = eig(S);
[~,idx] = sort(diag(L),'descend');
U = U(:,idx);
window = U(:,1:nTapers);
end
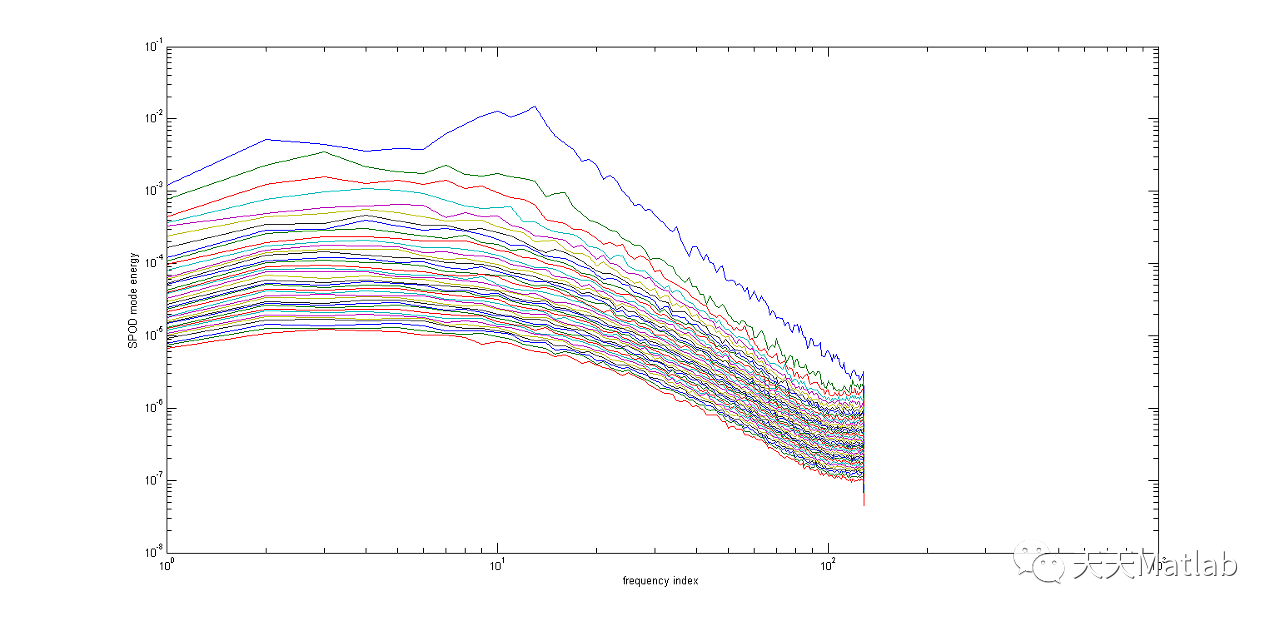
⛄ 运行结果
⛄ 参考文献
[1] A. Nekkanti, O. T. Schmidt, Frequency鈥搕ime analysis, low-rank reconstruction and denoising of turbulent flows using SPOD, Journal of Fluid Mechanics 926, A26, 2021
⛄ Matlab代码关注
❤️部分理论引用网络文献,若有侵权联系博主删除
❤️ 关注我领取海量matlab电子书和数学建模资料