kmeans++聚类生成anchors
说明
使用yolo系列通常需要通过kmeans聚类算法生成anchors,
但kmeans算法本身具有一定的局限性,聚类结果容易受初始值选取影响。
因此通过改进原kmeans_for_anchors.py实现 kmeans++聚类生成anchors。
具体实现如下:
import glob
import xml.etree.ElementTree as ET
from tqdm import tqdm
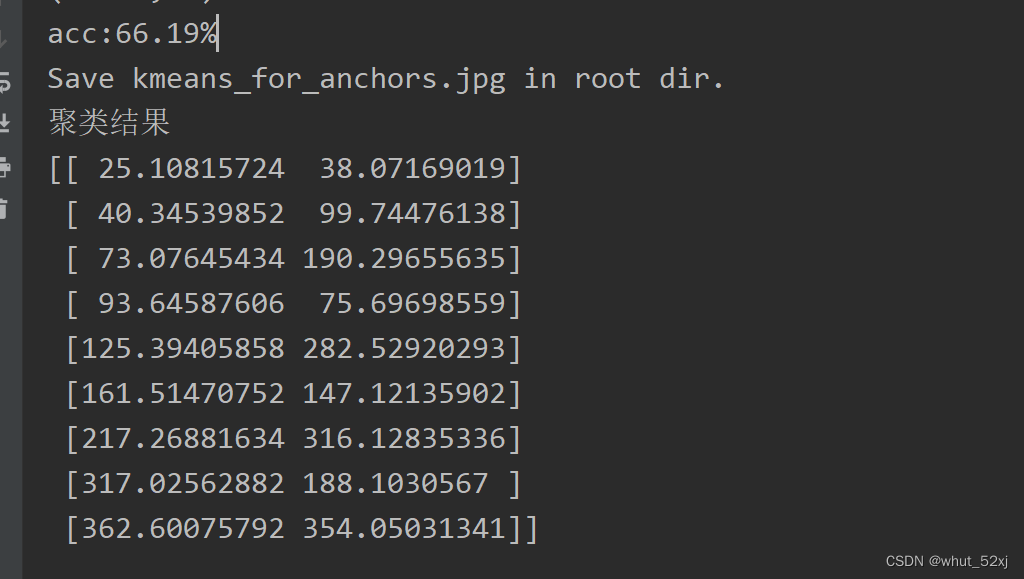
import numpy as npdef cas_iou(box, cluster):x = np.minimum(cluster[:, 0], box[0])y = np.minimum(cluster[:, 1], box[1])intersection = x * yarea1 = box[0] * box[1]area2 = cluster[:, 0] * cluster[:, 1]iou = intersection / (area1 + area2 - intersection)return ioudef avg_iou(box, cluster):return np.mean([np.max(cas_iou(box[i], cluster)) for i in range(box.shape[0])])def bboxesOverRation(bboxesA, bboxesB):"""功能等同于matlab的函数bboxesOverRationbboxesA:M*4 array,形如[x,y,w,h]排布bboxesB: N*4 array,形如[x,y,w,h]排布"""bboxesA = np.array(bboxesA.astype('float'))bboxesB = np.array(bboxesB.astype('float'))M = bboxesA.shape[0]N = bboxesB.shape[0]areasA = bboxesA[:, 2] * bboxesA[:, 3]areasB = bboxesB[:, 2] * bboxesB[:, 3]xA = bboxesA[:, 0] + bboxesA[:, 2]yA = bboxesA[:, 1] + bboxesA[:, 3]xyA = np.stack([xA, yA]).transpose()xyxyA = np.concatenate((bboxesA[:, :2], xyA), axis=1)xB = bboxesB[:, 0] + bboxesB[:, 2]yB = bboxesB[:, 1] + bboxesB[:, 3]xyB = np.stack([xB, yB]).transpose()xyxyB = np.concatenate((bboxesB[:, :2], xyB), axis=1)iouRatio = np.zeros((M, N))for i in range(M):for j in range(N):x1 = max(xyxyA[i, 0], xyxyB[j, 0]);x2 = min(xyxyA[i, 2], xyxyB[j, 2]);y1 = max(xyxyA[i, 1], xyxyB[j, 1]);y2 = min(xyxyA[i, 3], xyxyB[j, 3]);Intersection = max(0, (x2 - x1)) * max(0, (y2 - y1));Union = areasA[i] + areasB[j] - Intersection;iouRatio[i, j] = Intersection / Union;return iouRatiodef load_data(path):data = []# 对于每一个xml都寻找boxfor xml_file in tqdm(glob.glob('{}/*xml'.format(path))):tree = ET.parse(xml_file)height = int(tree.findtext('./size/height'))width = int(tree.findtext('./size/width'))if height <= 0 or width <= 0:continue# 对于每一个目标都获得它的宽高for obj in tree.iter('object'):xmin = int(float(obj.findtext('bndbox/xmin'))) / widthymin = int(float(obj.findtext('bndbox/ymin'))) / heightxmax = int(float(obj.findtext('bndbox/xmax'))) / widthymax = int(float(obj.findtext('bndbox/ymax'))) / heightxmin = np.float64(xmin)ymin = np.float64(ymin)xmax = np.float64(xmax)ymax = np.float64(ymax)# 得到宽高x = xmin + 0.5 * (xmax - xmin)y = ymin + 0.5 * (ymax - ymin)data.append([x, y, xmax - xmin, ymax - ymin])return np.array(data)def estimateAnchorBoxes(trainingData, numAnchors=9):'''功能:kmeans++算法估计anchor,类似于matlab函数estimateAnchorBoxes,当trainingData数据量较大时候,自写的kmeans迭代循环效率较低,matlab的estimateAnchorBoxes得出anchors较快,但meanIOU较低,然后乘以实际box的ratio即可。此算法由于优化是局部,易陷入局部最优解,结果不一致属正常cuixingxing150@gmail.comExample:import scipy.io as scipodata = scipo.loadmat(r'D:\Matlab_files\trainingData.mat')trainingData = data['temp']meanIoUList = []for numAnchor in np.arange(1,16):anchorBoxes,meanIoU = estimateAnchorBoxes(trainingData,numAnchors=numAnchor)meanIoUList.append(meanIoU)plt.plot(np.arange(1,16),meanIoUList,'ro-')plt.ylabel("Mean IoU")plt.xlabel("Number of Anchors")plt.title("Number of Anchors vs. Mean IoU")Parameters----------trainingData : numpy 类型形如[x,y,w,h]排布,M*4大小二维矩阵numAnchors : int, optional估计的anchors数量. The default is 9.Returns-------anchorBoxes : numpy类型形如[w,h]排布,N*2大小矩阵.meanIoU : scalar 标量DESCRIPTION.'''numsObver = trainingData.shape[0]xyArray = np.zeros((numsObver, 2))trainingData[:, 0:2] = xyArrayassert (numsObver >= numAnchors)# kmeans++# initcentroids = [] # 初始化中心,kmeans++centroid_index = np.random.choice(numsObver, 1)centroids.append(trainingData[centroid_index])while len(centroids) < numAnchors:minDistList = []for box in trainingData:box = box.reshape((-1, 4))minDist = 1for centroid in centroids:centroid = centroid.reshape((-1, 4))ratio = (1 - bboxesOverRation(box, centroid)).item()if ratio < minDist:minDist = ratiominDistList.append(minDist)sumDist = np.sum(minDistList)prob = minDistList / sumDistidx = np.random.choice(numsObver, 1, replace=True, p=prob)centroids.append(trainingData[idx])# kmeans 迭代聚类maxIterTimes = 100iter_times = 0while True:minDistList = []minDistList_ind = []for box in trainingData:box = box.reshape((-1, 4))minDist = 1box_belong = 0for i, centroid in enumerate(centroids):centroid = centroid.reshape((-1, 4))ratio = (1 - bboxesOverRation(box, centroid)).item()if ratio < minDist:minDist = ratiobox_belong = iminDistList.append(minDist)minDistList_ind.append(box_belong)centroids_avg = []for _ in range(numAnchors):centroids_avg.append([])for i, anchor_id in enumerate(minDistList_ind):centroids_avg[anchor_id].append(trainingData[i])err = 0for i in range(numAnchors):if len(centroids_avg[i]):temp = np.mean(centroids_avg[i], axis=0)err += np.sqrt(np.sum(np.power(temp - centroids[i], 2)))centroids[i] = np.mean(centroids_avg[i], axis=0)iter_times += 1if iter_times > maxIterTimes or err == 0:breakanchorBoxes = np.array([x[2:] for x in centroids])meanIoU = 1 - np.mean(minDistList)print('acc:{:.2f}%'.format(avg_iou(trainingData[:, 2:], anchorBoxes) * 100))return anchorBoxes, meanIoUif __name__ == "__main__":np.random.seed(0)# 载入数据集,可以使用VOC的xmlpath = 'VOCdevkit/VOC2007/Annotations'# 生成的anchors的txt文件保存路径anchorsPath = 'yolo_anchors++.txt'# 生成的anchors数量anchors_num = 9# 输入的图片尺寸input_shape = [416, 416]print('Load xmls.')data = load_data(path)print('Load xmls done.')# 使用k聚类算法print('K-means++ boxes.')anchors, _= estimateAnchorBoxes(data, numAnchors=anchors_num)print('K-means boxes done.')anchors = anchors * np.array([input_shape[1], input_shape[0]])# 排序cluster = anchors[np.argsort(anchors[:, 0])]print("聚类结果")print(cluster)# 保存结果 生成yolo_anchors++.txt文件f = open(anchorsPath, 'w')row = np.shape(cluster)[0]for i in range(row):if i == 0:x_y = "%d,%d" % (cluster[i][0], cluster[i][1])else:x_y = ", %d,%d" % (cluster[i][0], cluster[i][1])f.write(x_y)f.close()使用voc数据集运行结果如下:
代码运行时间比较长,耐心等待即可!
参考
1.YOLOV4生成锚框kmeans_for_anchors.py
#-------------------------------------------------------------------------------------------------------#
# kmeans虽然会对数据集中的框进行聚类,但是很多数据集由于框的大小相近,聚类出来的9个框相差不大,
# 这样的框反而不利于模型的训练。因为不同的特征层适合不同大小的先验框,shape越小的特征层适合越大的先验框
# 原始网络的先验框已经按大中小比例分配好了,不进行聚类也会有非常好的效果。
#-------------------------------------------------------------------------------------------------------#
import glob
import xml.etree.ElementTree as ETimport matplotlib.pyplot as plt
import numpy as np
from tqdm import tqdmdef cas_iou(box, cluster):x = np.minimum(cluster[:, 0], box[0])y = np.minimum(cluster[:, 1], box[1])intersection = x * yarea1 = box[0] * box[1]area2 = cluster[:,0] * cluster[:,1]iou = intersection / (area1 + area2 - intersection)return ioudef avg_iou(box, cluster):return np.mean([np.max(cas_iou(box[i], cluster)) for i in range(box.shape[0])])def kmeans(box, k):#-------------------------------------------------------------## 取出一共有多少框#-------------------------------------------------------------#row = box.shape[0]#-------------------------------------------------------------## 每个框各个点的位置#-------------------------------------------------------------#distance = np.empty((row, k))#-------------------------------------------------------------## 最后的聚类位置#-------------------------------------------------------------#last_clu = np.zeros((row, ))np.random.seed()#-------------------------------------------------------------## 随机选5个当聚类中心#-------------------------------------------------------------#cluster = box[np.random.choice(row, k, replace = False)]iter = 0while True:#-------------------------------------------------------------## 计算当前框和先验框的宽高比例#-------------------------------------------------------------#for i in range(row):distance[i] = 1 - cas_iou(box[i], cluster)#-------------------------------------------------------------## 取出最小点#-------------------------------------------------------------#near = np.argmin(distance, axis=1)if (last_clu == near).all():break#-------------------------------------------------------------## 求每一个类的中位点#-------------------------------------------------------------#for j in range(k):cluster[j] = np.median(box[near == j],axis=0)last_clu = nearif iter % 5 == 0:print('iter: {:d}. avg_iou:{:.2f}'.format(iter, avg_iou(box, cluster)))iter += 1return cluster, neardef load_data(path):data = []#-------------------------------------------------------------## 对于每一个xml都寻找box#-------------------------------------------------------------#for xml_file in tqdm(glob.glob('{}/*xml'.format(path))):tree = ET.parse(xml_file)height = int(tree.findtext('./size/height'))width = int(tree.findtext('./size/width'))if height<=0 or width<=0:continue#-------------------------------------------------------------## 对于每一个目标都获得它的宽高#-------------------------------------------------------------#for obj in tree.iter('object'):xmin = int(float(obj.findtext('bndbox/xmin'))) / widthymin = int(float(obj.findtext('bndbox/ymin'))) / heightxmax = int(float(obj.findtext('bndbox/xmax'))) / widthymax = int(float(obj.findtext('bndbox/ymax'))) / heightxmin = np.float64(xmin)ymin = np.float64(ymin)xmax = np.float64(xmax)ymax = np.float64(ymax)# 得到宽高data.append([xmax - xmin, ymax - ymin])return np.array(data)if __name__ == '__main__':np.random.seed(0)#-------------------------------------------------------------## 运行该程序会计算'./VOCdevkit/VOC2007/Annotations'的xml# 会生成yolo_anchors.txt#-------------------------------------------------------------#input_shape = [416, 416]anchors_num = 9#-------------------------------------------------------------## 载入数据集,可以使用VOC的xml#-------------------------------------------------------------#path = 'C:\\Users\\52xj\\Desktop\\mobilenet-yolov4-pytorch-main\\mobilenet-yolov4-pytorch-main\\VOCdevkit\\VOC2007\\Annotations'#-------------------------------------------------------------## 载入所有的xml# 存储格式为转化为比例后的width,height#-------------------------------------------------------------#print('Load xmls.')data = load_data(path)print(data.shape)data = data * np.array([input_shape[1], input_shape[0]])print(data.shape)print('Load xmls done.')#-------------------------------------------------------------## 使用k聚类算法#-------------------------------------------------------------#print('K-means boxes.')cluster, near = kmeans(data, anchors_num)print('K-means boxes done.')data = data * np.array([input_shape[1], input_shape[0]])print(data.shape)cluster = cluster * np.array([input_shape[1], input_shape[0]])#-------------------------------------------------------------## 绘图#-------------------------------------------------------------#for j in range(anchors_num):plt.scatter(data[near == j][:,0], data[near == j][:,1])plt.scatter(cluster[j][0], cluster[j][1], marker='x', c='black')plt.savefig("kmeans_for_anchors.jpg")plt.show()print('Save kmeans_for_anchors.jpg in root dir.')cluster = cluster[np.argsort(cluster[:, 0] * cluster[:, 1])]print('avg_ratio:{:.2f}'.format(avg_iou(data, cluster)))print(cluster)f = open("yolo_anchors.txt", 'w')row = np.shape(cluster)[0]for i in range(row):if i == 0:x_y = "%d,%d" % (cluster[i][0], cluster[i][1])else:x_y = ", %d,%d" % (cluster[i][0], cluster[i][1])f.write(x_y)f.close()2.Kmeans++聚类算法
重新思考Anchor Box估计

代码实现
def bboxesOverRation(bboxesA,bboxesB):"""功能等同于matlab的函数bboxesOverRationbboxesA:M*4 array,形如[x,y,w,h]排布bboxesB: N*4 array,形如[x,y,w,h]排布"""bboxesA = np.array(bboxesA.astype('float'))bboxesB = np.array(bboxesB.astype('float'))M = bboxesA.shape[0]N = bboxesB.shape[0]areasA = bboxesA[:,2]*bboxesA[:,3]areasB = bboxesB[:,2]*bboxesB[:,3]xA = bboxesA[:,0]+bboxesA[:,2]yA = bboxesA[:,1]+bboxesA[:,3]xyA = np.stack([xA,yA]).transpose()xyxyA = np.concatenate((bboxesA[:,:2],xyA),axis=1)xB = bboxesB[:,0] +bboxesB[:,2]yB = bboxesB[:,1]+bboxesB[:,3]xyB = np.stack([xB,yB]).transpose()xyxyB = np.concatenate((bboxesB[:,:2],xyB),axis=1)iouRatio = np.zeros((M,N))for i in range(M):for j in range(N):x1 = max(xyxyA[i,0],xyxyB[j,0]);x2 = min(xyxyA[i,2],xyxyB[j,2]);y1 = max(xyxyA[i,1],xyxyB[j,1]);y2 = min(xyxyA[i,3],xyxyB[j,3]);Intersection = max(0,(x2-x1))*max(0,(y2-y1));Union = areasA[i]+areasB[j]-Intersection;iouRatio[i,j] = Intersection/Union; return iouRatiodef estimateAnchorBoxes(trainingData,numAnchors=9):'''功能:kmeans++算法估计anchor,类似于matlab函数estimateAnchorBoxes,当trainingData数据量较大时候,自写的kmeans迭代循环效率较低,matlab的estimateAnchorBoxes得出anchors较快,但meanIOU较低,然后乘以实际box的ratio即可。此算法由于优化是局部,易陷入局部最优解,结果不一致属正常cuixingxing150@gmail.comExample: import scipy.io as scipodata = scipo.loadmat(r'D:\Matlab_files\trainingData.mat')trainingData = data['temp']meanIoUList = []for numAnchor in np.arange(1,16):anchorBoxes,meanIoU = estimateAnchorBoxes(trainingData,numAnchors=numAnchor)meanIoUList.append(meanIoU)plt.plot(np.arange(1,16),meanIoUList,'ro-')plt.ylabel("Mean IoU")plt.xlabel("Number of Anchors")plt.title("Number of Anchors vs. Mean IoU")Parameters----------trainingData : numpy 类型形如[x,y,w,h]排布,M*4大小二维矩阵numAnchors : int, optional估计的anchors数量. The default is 9.Returns-------anchorBoxes : numpy类型形如[w,h]排布,N*2大小矩阵.meanIoU : scalar 标量DESCRIPTION.'''numsObver = trainingData.shape[0]xyArray = np.zeros((numsObver,2))trainingData[:,0:2] = xyArrayassert(numsObver>=numAnchors)# kmeans++# init centroids = [] # 初始化中心,kmeans++centroid_index = np.random.choice(numsObver, 1)centroids.append(trainingData[centroid_index])while len(centroids)<numAnchors:minDistList = []for box in trainingData:box = box.reshape((-1,4))minDist = 1for centroid in centroids:centroid = centroid.reshape((-1,4))ratio = (1-bboxesOverRation(box,centroid)).item()if ratio<minDist:minDist = ratiominDistList.append(minDist)sumDist = np.sum(minDistList)prob = minDistList/sumDist idx = np.random.choice(numsObver,1,replace=True,p=prob)centroids.append(trainingData[idx])# kmeans 迭代聚类maxIterTimes = 100iter_times = 0while True:minDistList = []minDistList_ind = []for box in trainingData:box = box.reshape((-1,4))minDist = 1box_belong = 0for i,centroid in enumerate(centroids):centroid = centroid.reshape((-1,4))ratio = (1-bboxesOverRation(box,centroid)).item()if ratio<minDist:minDist = ratiobox_belong = iminDistList.append(minDist)minDistList_ind.append(box_belong)centroids_avg = []for _ in range(numAnchors):centroids_avg.append([])for i,anchor_id in enumerate(minDistList_ind):centroids_avg[anchor_id].append(trainingData[i])err = 0for i in range(numAnchors):if len(centroids_avg[i]):temp = np.mean(centroids_avg[i],axis=0)err += np.sqrt(np.sum(np.power(temp-centroids[i],2)))centroids[i] = np.mean(centroids_avg[i],axis=0)iter_times+=1if iter_times>maxIterTimes or err==0:breakanchorBoxes = np.array([x[2:] for x in centroids])meanIoU = 1-np.mean(minDistList)return anchorBoxes,meanIoU